Move 78: Toward a Post Human Intelligence
2021/11/12
2021/11/12 Deep learning methods for designing proteins scaffolding functional sites, by Jue Wang, Sidney Lisanza, David Juergens, Doug Tischer, Ivan Anishchenko, Minkyung Baek, Joseph L Watson, Jung-Ho Chun, Lukas F. Milles, Justas Dauparas, Marc Expòsit, Wei Yang, Amijai Saragovi, Sergey Ovchinnikov, David Baker. bioRxiv
Abstract: Current approaches to de novo design of proteins harboring a desired binding or catalytic motif require pre-specification of an overall fold or secondary structure composition, and hence considerable trial and error can be required to identify protein structures capable of scaffolding an arbitrary functional site. Here we describe two complementary approaches to the general functional site design problem that employ the RosettaFold and AlphaFold neural networks which map input sequences to predicted structures. ... We show that the two approaches have considerable synergy, and AlphaFold2 structure prediction calculations suggest that the approaches can accurately generate proteins containing a very wide array of functional sites.
Download the pdf or the supplementary material
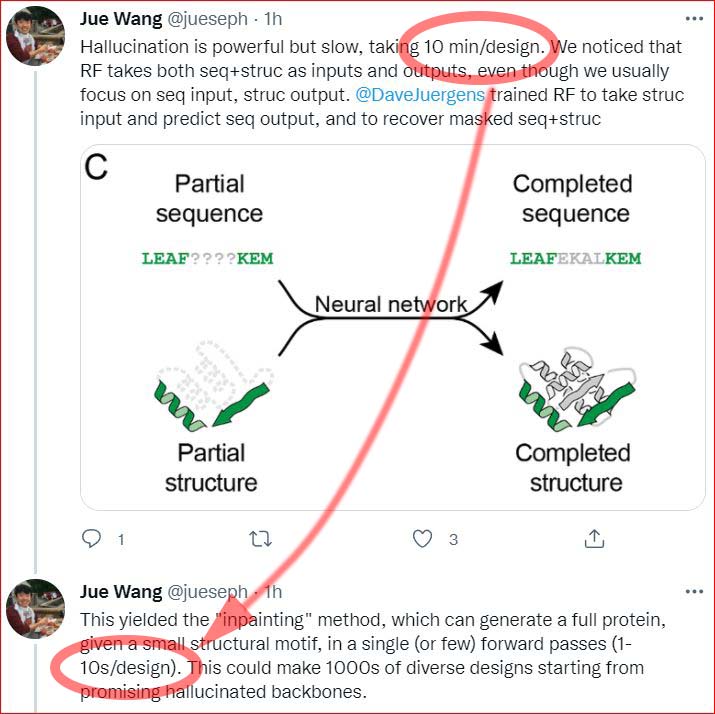
The above paper's predecessor: 2020/11/29 Design of proteins presenting discontinuous functional sites using deep learning, by Doug Tischer, Sidney Lisanza, Jue Wang, Runze Dong, Ivan Anishchenko, Lukas F. Milles, Sergey Ovchinnikov, and David Baker. bioRxiv
Abstract: An outstanding challenge in protein design is the design of binders against therapeutically relevant target proteins via scaffolding the discontinuous binding interfaces present in their often large and complex binding partners. There is currently no method for sampling through the almost unlimited number of possible protein structures for those capable of scaffolding a specified discontinuous functional site; instead, current approaches make the sampling problem tractable by restricting search to structures composed of pre-defined secondary structural elements. Such restriction of search has the disadvantage that considerable trial and error can be required to identify architectures capable of scaffolding an arbitrary discontinuous functional site, and only a tiny fraction of possible architectures can be explored. Here we build on recent advances in de novo protein design by deep network hallucination to develop a solution to this problem which eliminates the need to pre-specify the structure of the scaffolding in any way. We use the trRosetta residual neural network, which maps input sequences to predicted inter-residue distances and orientations, to compute a loss function which simultaneously rewards recapitulation of a desired structural motif and the ideality of the surrounding scaffold, and generate diverse structures harboring the desired binding interface by optimizing this loss function by gradient descent. We illustrate the power and versatility of the method by scaffolding binding sites from proteins involved in key signaling pathways with a wide range of secondary structure compositions and geometries. The method should be broadly useful for designing small stable proteins containing complex functional sites.
(2021/7/22) Deep Mind AlphaFold Predicts the Shapes and 3-D structure of 350,000 proteins, including every one made by humans, by Cade Metz, The New York Times
On Thursday, DeepMind released the predicted shapes of more than 350,000 proteins — the microscopic mechanisms that drive the behavior of bacteria, viruses, the human body and all other living things. This new database includes the three-dimensional structures for all proteins expressed by the human genome, as well as those for proteins that appear in 20 other organisms, including the mouse, the fruit fly and the E. coli bacterium.
“I thought it would take another 10 years,” said Randy Read, a professor at the University of Cambridge. “This was a complete change.”
This new knowledge is its own sort of key: If scientists can determine the shape of a protein, they can determine how other molecules will bind to it. This might reveal, say, how bacteria resist antibiotics — and how to counter that resistance. Bacteria resist antibiotics by expressing certain proteins; if scientists were able to identify the shapes of these proteins, they could develop new antibiotics or new medicines that suppress them.
The system’s accuracy does vary, so some of the predictions in DeepMind’s database will be less useful than others. Each prediction in the database comes with a “confidence score” indicating how accurate it is likely to be. DeepMind researchers estimate that the system provides a “good” prediction about 95 percent of the time.
2021/10/13 Artificial insect-inspired ‘brain’ can guide robotic dog through maze, by Edd Gent, NewScientist
The control system, built by UK start-up company Opteran Technologies, mimics how honeybees and other insects navigate. It can be connected to various robots and drones. The package weighs only 30 grams and draws less than 3 watts of electricity.
2021/05/26 Panorama - Are You Scared Yet, Human? BBC [You'll need a VPN to access. DRM restricts this video to U.K. play]
From Amazon’s Alexa to improvements in cancer care, artificial intelligence is changing our world. But today leading tech figures from Silicon Valley worry about the future that’s being created. Brad Smith, president of Microsoft, believes George Orwell's 1984 could become reality by 2024. Panorama has uncovered new evidence of AI being used by police in China to recognise the emotions of detainees in order to help determine guilt or innocence. China has vowed to become the world's AI superpower by 2030, sparking a new arms race with America. Both countries are pouring billions into cutting-edge military tech, including autonomous weapons. AI could usher in a golden age, but without urgent regulation, experts warn we could lose control of artificial intelligence, a prospect, they say, that should scare us all.
2021/10/06 A census of cell types in the brain’s motor cortex, by Johan Winnubst & Silvia Arber, Nature
2020/02/24 A decade of questions about the fluidity of cell identity, by Giacomo Masserdotti & Magdalena Götz, Nature
A method for directly converting connective-tissue cells into neurons opened up a new branch of research into cell-based therapies and called into question long-held beliefs about how development affects a cell’s identity.
2010/02/24 Cell reprogramming gets direct, by Cory R. Nicholas & Arnold R. Kriegstein, Nature
In a feat of biological wizardry, one type of differentiated cell has been directly converted into another, completely distinct type. Notably, the approach does not require a stem-cell intermediate stage.
2010/01/27 Direct conversion of fibroblasts to functional neurons by defined factors, by Thomas Vierbuchen, Austin Ostermeier, Zhiping P. Pang, Yuko Kokubu, Thomas C. Südhof & Marius Wernig, Nature
Cellular differentiation and lineage commitment are considered to be robust and irreversible processes during development. Recent work has shown that mouse and human fibroblasts can be reprogrammed to a pluripotent state with a combination of four transcription factors. This raised the question of whether transcription factors could directly induce other defined somatic cell fates, and not only an undifferentiated state. We hypothesized that combinatorial expression of neural-lineage-specific transcription factors could directly convert fibroblasts into neurons.
2019/08/21 A recipe book for cell types in the human brain, by Matthew G. Keefe & Tomasz J. Nowakowski, Natureee
Whether cell types in the brain have been conserved during evolution is not clear. A comparison of the molecular recipes that define brain cell types in humans and mice reveals similarities and differences between species.
2019/08/21 Conserved cell types with divergent features in human versus mouse cortex, by Rebecca D. Hodge, Trygve E. Bakken, […]Ed S. Lein, Nature
2018/10/31 Single-cell sequencing paints diverse pictures of the brain, by Aparna Bhaduri & Tomasz J. Nowakowski, Nature
A single-cell sequencing study reveals how different types of neuron are distributed in the brain. An analysis then demonstrates how these data can improve our understanding of neuronal functions.
1/16/2020 Why did Lee Sedol, one of the world’s best ‘Go’ players, retire from the game? TRT
One of the greatest ‘Go’ players of all time just retired from the game, saying that artificial intelligence programs can play at a level humans aren’t able to. His decision has reignited discussions about AI and how we define human achievement.
AlphaGo
DeepMind’s Demis Hassabis on its breakthrough scientific discoveries | WIRED Live, 3/5/2021
David Silver: AlphaGo, AlphaZero, and Deep Reinforcement Learning | Lex Fridman Podcast #86 (2020-04-03)
AlphaFold and the Grand Challenge to solve protein folding, Arxiv Insights, Youtube (7/27/2021)
AlphaFold: The making of a scientific breakthrough, by DeepMind, 11/30/2020
5 challenges we could solve by designing new proteins , by David Baker, 7/16/2019
The protein folding problem: a major conundrum of science: Ken Dill at TEDxSBU, 10/22/2013
Astonishing molecular machines: Drew Berry at TEDxSydney, 6/11/2011
RL Course by David Silver - Lecture 1: Introduction to Reinforcement Learning (2015-05-13)
RL Course by David Silver - Lecture 5: Model Free Control
RL Course by David Silver - Lecture 7: Policy Gradient Methods
Elliot Waite: Policy Gradient Theorem Explained - Reinforcement Learning (2020-11-22)
Alexander Amini: MIT 6.S191: Lecture 1, Foundations of Deep Learning (2021-03-05) Introduction to Deep Learning
Alexander Amini: MIT 6.S191: Lecture 1, Foundations of Deep Learning (2020-01-27) Introduction to Deep Learning
Lex Fridman: MIT 6.S091: Introduction to Deep Reinforcement Learning (Deep RL) (2019-01-24)
Ava Soleimany: MIT 6.S191: Lecture 2, Recurrent Neural Networks (2021-01-28) Introduction to Deep Learning
Alexander Amini: MIT 6.S191: Lecture 3, Convolutional Neural Networks (2021-01-20) Introduction to Deep Learning
Ava Soleimany: MIT 6.S191: Lecture 4, Deep Generative Modeling (2021-01-21) Introduction to Deep Learning
Alexander Amini: MIT 6.S191: Lecture 5, Deep Reinforcement Learning (2019-01-30) Introduction to Deep Learning
Alexander Amini: MIT 6.S191: Lecture 5, Deep Reinforcement Learning (2021-03-05) Introduction to Deep Learning
Ava Soleilmany: MIT 6.S191: Lecture 6, Deep Learning Limitations and New Frontiers (2021-03-12) Introduction to Deep Learning
Fernanda Viégas: MIT 6.S191, Lecture 7, Data Visualization for Machine Learning (2019-01-31) Introduction to Deep Learning
Alexander Amini: MIT 6.S191, Lecture 7, Evidential Deep Learning and Uncertainty Estimation (2021-01-26) Introduction to Deep Learning
Ava Soleimany: MIT 6.S191: Lecture 8, AI Bias and Fairness (2021-01-26) Introduction to Deep Learning
Prof. Sanja Fidler, NVIDIA: MIT 6.S191, Lecture 10, Toward AI for 3D Content Creation (2021-01-28) Introduction to Deep Learning
freeCodeCamp.org: Reinforcement Learning Course - Full Machine Learning Tutorial (2019-05-14)
ACMSIGGRAPH Deep Learning: A Crash Course (2018-08-12)
================================ Protein Folding ================================
[Fix the link]
ChimeraX @UCSFChimeraX
·
2021-09-09 The ChimeraX command to show these large AlphaFold models is here if you want to make them yourself.